CANBIO2 – Projects
Individual projects
Bitte beachten Sie, dass der Inhalt nur auf Englisch verfügbar ist.
WP1 – Novel Cancer Models
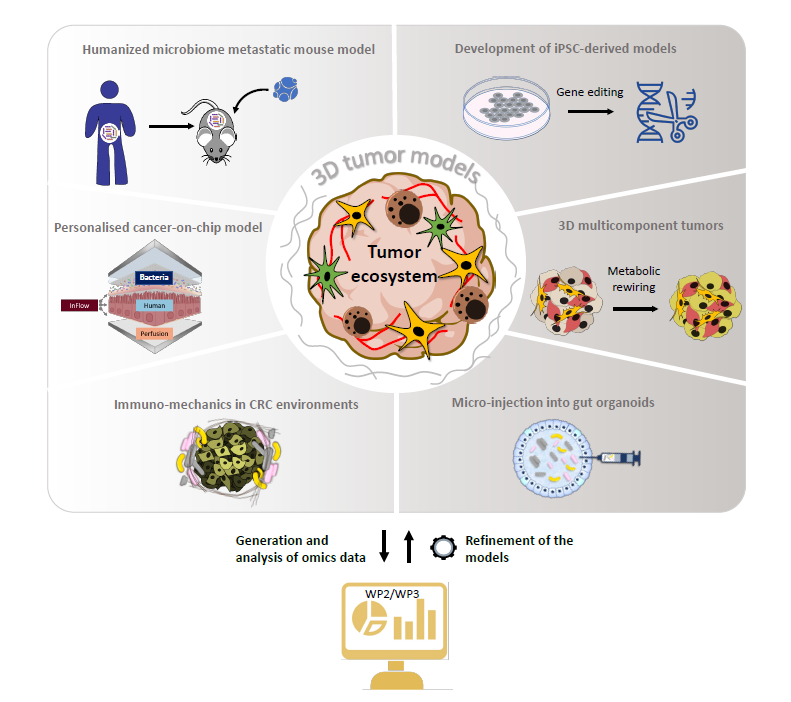
Project 1: Prof Stephanie Kreis (UL, Department of Life Sciences and Medicine, Esch-sur-Alzette): The MEL-MOD models: 3D multicomponent melanoma models as preclinical surrogates for drug and metabolic biomarker screening and assessment.
Recruitment closed. Position has been filled.
The MEL-MOD models are 3D multi-component melanoma spheroid models, where diverse cells of the TE (fibroblasts, immune cells and endothelial cells) are combined with tumour cells. These functionally and transcriptionally fully characterised different 3D cancer models will be used to study metabolic reprogramming in tumour cells and cells of the TE (P8, P10), as well as to identify new targets based on the TE-tumour cell interaction (WP3).

Prof Stephanie Kreis is Associate Professor at the Department of Life Sciences and Medicine where she co-heads the Signal Transduction Research group. Her current research interests center around drug resistance mechanisms in melanoma and developing sophisticated 3D models for high throughput drug screening and mechanistic studies. Prof Kreis is lecturing Biology Bachelor, Master as well as medical students. Supervising PhD students, she has developed a keen interest to ensure scientific and institutional quality of PhD education and is the acting Head of the UL Doctoral School for Science and Engineering.
Project 3: Supervisor Dr Clément Thomas (LIH, Department of Cancer Research, Luxembourg): Reciprocal effects of intrinsic tumour immune resistance and the tumour microenvironment.
Recruitment closed. Position has been filled.
The immune system is responsible for detecting and eliminating cancer cells. To kill their targets, cytotoxic lymphocytes use a specialised cell-to-cell interface, termed the immunological synapse (IS). Recently, our group established that cancer cells can escape from cytotoxic lymphocyte-mediated killing by polarizing their actin cytoskeleton to the IS and thereby altering IS morphology and activity. Actin-driven immune escape will be investigated in established 3D brain models including the immune compartment. Factors of the TE, such as hypoxia and ROS, will be monitored and their influence on actin dynamics in cancer cells and immune evasion will be determined.

Dr Clément Thomas’ research is centred on the functions and regulation of the actin cytoskeleton I in cancer cells, with a particular interest in actin-driven processes underlying tumour immune evasion and metastasis. In a recent study, his group established that a particular cytoskeletal structure that forms in cancer cells attacked by cytotoxic lymphocytes represents a major point of convergence for multiple immune evasion mechanisms and a promising therapeutic point of intervention to restore an effective anti-tumour immune response in patients.
Project 4: Supervisor Prof Lasse Sinkkonen (UL, Department of Life Sciences and Medicine, Esch-sur-Alzette): Epigenomic Response of Colon Cancer Cells to Fusobacterium nucleatum Exposure.
Recruitment closed. Position has been filled.
Micro-injection of single bacteria into a gut organoid model has recently been used to reveal the causative role of a genotoxic pks+E.coli mutational signature in CRC. Our team is currently establishing micro-injection of the well described CRC-associated bacteria, Fusobacterium nucleatum (to be extended during the course of the project to complex microbiome communities), into tumour organoid and preliminary data suggest extensive epigenetic remodelling in tumour cells. We will further use this micro-injection-based model to determine chromatin accessibility by single nuclei ATAC-seq upon single or complex microbiota challenge of tumour cells, thereby distinguishing between changes induced by microbiota secretome and direct microbe interactions.

Prof Lasse Sinkkonen holds a PhD in Biochemistry from University of Basel and is currently the Head of the Epigenetics team at the UL Department of Life Sciences and Medicine. His research is focused on understanding the gene regulatory control of cell identity at the level of chromatin in both development and disease. The Epigenetics team applies genome-wide and single cell technologies to reveal the key regulators of cell states and the associated cellular heterogeneity during differentiation and in diseases such as Parkinson’s disease and cancer. A particular focus is placed on the interplay between metabolites and epigenetic control of gene expression in cancer.
Project 5: Supervisor Prof Anupam Sengupta (UL, Department of Physics and Materials Science, Luxembourg): Active Immuno-Mechanics in CRC environments (AIM-CRC).
Recruitment closed. Position has been filled.
The role of host feedback on the microbial community has been largely neglected until now. Our team has developed diverse workflows, which allow to track microbial behaviour using microfluidics, advanced imaging, in-house-robotics, mathematics and machine learning tools. Here we will use these tools and combine physics, mathematical sciences, and biology to understand how biophysical parameters (ECM, rigidity) within the TE regulate the dynamics of the cancer microbiome. The advantage of such models is that the cell-to-cell and cell-to-matrix interactions, which are essential for mimicking accurate physiological conditions and responses, are maintained.

Prof Anupam Sengupta is an FNR ATTRACT Fellow and Head of the Physics of Living Matter group at UL Department of Physics and Materials Science. After obtaining BS-MS degrees in Mechanical Engineering from the IIT Bombay (India), Anupam joined the Max Planck Institute (Göttingen, Germany). In 2013 he received a PhD in Physics for his work on liquid crystal microfluidics. Prof Sengupta was a Human Frontier Cross-Disciplinary Fellow (2014-2017), first at the MIT (USA) and then at ETH Zurich (Switzerland), working on the physical ecology of microbes. Since 2018, he directs a multi-disciplinary team in Luxembourg, combining material physics, microbiology, mathematical modelling and machine learning to understand microbial response and adaptation. He is a member of the UL Institute of Advanced Studies and serves as the Director of the undergraduate physics studies.
Project 6: Supervisor Prof Paul Wilmes (UL, Luxembourg Centre for Systems Biomedicine, Esch-sur-Alzette): A personalised cancer-on-chip model for the study of complex gut microbiota in colorectal cancer initiation and progression.
Recruitment closed. Position has been filled.
Recently, it has become increasingly evident that focus should not only be put on individual bacterial species, but also on the various metabolites that they secrete. The microfluidics HuMiX device, developed at UL, was already used within the consortium to study single bacteria-host interaction with tumour and immune cells. In CANBIO2, it will be adapted and transformed into a cancer-on-a-chip model to study the metabolic cross-talk taking place between complex patient-derived microbiome communities and host cells. The outcome of these projects will lead to the identification of metabolites that can be specifically studied in WP2 (P8, P10).

Prof Paul Wilmes is Professor of Systems Ecology at the UL. As a British Chevening Scholar, Paul earned his PhD from the School of Environmental Sciences at the University of East Anglia (UK). For part of his doctoral research, he spent time as a German Academic Exchange Service Visiting Scientist at the Max Planck Institute for Marine Microbiology in Bremen (Germany). Paul subsequently carried out postdoctoral research at the University of California, Berkeley (USA) from where he returned in 2010 to his native Luxembourg through the FNR ATTRACT fellowship scheme Paul was awarded an European Research Council (ERC) Consolidator Grant in 2019.
Project 7: Supervisor Dr Elisabeth Letellier – DTU co-coordinator (UL, Department of Life Sciences and Medicine, Esch-sur-Alzette): A humanised microbiome metastatic mouse model to develop microbiome-based therapeutic strategies.
Recruitment closed. Position has been filled.
Our team has recently established a humanised microbiome model (based on faecal transplantation of human microbiota in germ free mice performed in our recently established germ-free mouse facility). This avatar model will now be further developed and coupled to a colonoscopy-based orthotopic injection of organoids in the colon wall to recapitulate the full metastatic cascade. They will help us determine the role of the microbiota in metastases and identify new strategies to reverse the harmful effects of cancer-associated bacteria. Importantly, these avatar models can be further extended to other tumour types and are at the disposal of all the projects within the consortium to validate microbiome-based targets.

Dr Elisabeth Letellier is co-heading the Molecular Disease Mechanisms group at the UL Department of Life Sciences and Medicine. Her current research activities mainly aim at understanding the mechanisms underlying tumour initiation and progression in colorectal cancer (CRC), with a special focus on the tumour microenvironment (TME). The group has developed state-of-the-art in silico (different bioinformatics tools as well as metabolic modelling), in vitro (3D organoids cultures of patient-derived samples) and in vivo models (CRC models and gnotobiotic mice) to understand how the different cell types of the TME, including immune cells and the cancer-associated fibroblasts as well as the different bacterial species of the gut microbiome, interact with the tumour cells.

discover CANBIO2
Projects – WP1 – Novel Cancer Models
Projects – WP2 – Cancer Metabolism
Projects – WP3 – Novel Treatment Strategies
Contact
For any question related to CANBIO2, please contact: